Nucleotide Repetition Lengths
code clojure
Sunday, November 3, 2013
After a long hiatus, let’s continue our foray into amateur genomics with Clojure, in which we continue to encounter elegant expressions of Clojure’s power as well as a few additional wrinkles in the Clojure ecosystem (in this case, Incanter).
In our last post we upgraded our genome decoder and used it to show frequencies of the four nucleotides, illustrating Chargaff’s Rule.
Another simple question to be asked of the data is, What is the
distribution of nucleotide repetition lengths? In other words, if we
represent the number of times any nucleotide is repeated as a
histogram, then AAGGCATTTT would yield two entries for 1, two entries
for 2, zero for 3, and one for 4. What does this distribution look
like for the entire human or yeast genome?
As a “null hypothesis”, consider the likelihood of getting \(N\) A’s when drawing at random from A, G, C or T. The probability for the first selection to be A is 25%; for the first two draws to be AA the probability is \(P(AA) = {1 \over {4^2}} = 0.625\), and for \(N\) consecutive draws, \(P(A \times N) = 4^{-N}\).
You can easily simulate (“Monto Carlo”) such a situation using a purely random sequence:
(defn randgenome [] (repeatedly #(rand-nth [:A :G :C :T])))
This function yields a sequence like (:A :A :C :A :T :G :G :T :G :A :C :C ...).
To convert this into repetition lengths, we can first divide the
sequence up into repeating elements using partition-by and the
identity function:
(partition-by identity (randgenome)) ;;=> ((:A :A) (:C) (:A) (:T) (:G :G) (:T) (:G) (:A) (:C :C) ...)
Extracting the lengths is then trivial: just map count over the
sequences:
(defn get-lengths [s]
(->> s
(partition-by identity)
(map count)))
(get-lengths (randgenome))
;;=> (2 1 1 1 2 1 1 1 2 ...)
(Note: running these functions unmodified will hang your REPL, since
randgenome yields an infinite sequence! Wrap them with take if
needed.)
Histogramming the data
The plots in my previous posts were made by the simple easy
expedient of copying data into a Numbers spreadsheet on my Mac. We’d
like to eventually do some more ambitious plotting, however, so let’s
investigate a Clojure-based solution.
Incanter provides a broad set of Clojure-based utilities for data analysis, including matrices and linear algebra operations, statistical distributions, and so on. Built on top of JFreeChart, it implements several different kinds of plots, including histograms, and it should be an obvious candidate for displaying our length distribution histogram.
Unfortunately, because of two issues I discovered, Incanter will not work for our purposes. To make a long story short, it does not handle empty (zero) entries in histograms properly when displaying on a logarithmic scale, and it generates out-of-memory errors when given more than a million or so entries. (Coming from high energy physics, where such histograms are extremely common, these both seem like serious flaws to me, which I hope will be remedied in the next Incanter release.)
I wound up using the JFreeChart API directly, though this was very time consuming, as the API has approximately the same complexity as the entire US space program or the Large Hadron Collider. (In what follows below I elide details about false starts, descriptions of hair pulling, and expletives.)
First, I needed to convert the lengths from get-lengths into a
histogram with arbitrarily many entries:
(defn make-hist
"
Convert seq of input xs into a histogram of nbins bins, from xmin to
xmax. Discard overflows or underflows
"
[xmin xmax nbins xs]
(let [;; "base" histogram (zeros):
zero-map (into (sorted-map)
(map (fn [x] [x 0]) (range nbins)))
;; get actual bin values for every input in xs:
xbins (map #(int (* nbins (/ (- % xmin)
(- xmax xmin))))
xs)
;; strip out undeflows & overflows:
no-overflows (->> xbins
(remove #(< % 0))
(remove #(>= % nbins)))]
;; yield histogram as array of [ibin, height] pairs:
(into [] (reduce #(update-in %1 [%2] inc) zero-map no-overflows))))
Supplying the minimum and maximum value ahead of time simplifies the histogramming code substantially and allows us to choose the same range for multiple plots, as will be shown below.
Armed with this transformation, the generation of the histogram plot is as follows:
(ns jenome.graphs
(:import [org.jfree.data.xy XYSeriesCollection XYSeries]
[org.jfree.chart ChartFrame JFreeChart]
[org.jfree.chart.plot XYPlot]
[org.jfree.chart.axis NumberAxis]
[org.jfree.chart.renderer.xy XYBarRenderer StandardXYBarPainter]
[org.jfree.chart.renderer.category]))
(defn trim-zeros
"
Convert zeros (or negatives) to small positive values to allow for
graphing on log scale
"
[vals]
(map (fn [[x y]] [x (if (> y 0) y 0.0001)]) vals))
(defn draw-hist
"
Draw histogram of bins as generated by make-hist
"
[x-label values]
(let [renderer (XYBarRenderer.)
painter (StandardXYBarPainter.)
series (XYSeries. [])
blue (java.awt.Color. 0x3b 0x6c 0x9d)
coll (XYSeriesCollection. series)
y-axis (org.jfree.chart.axis.LogarithmicAxis. "Entries")
plot (XYPlot. coll (NumberAxis. x-label) y-axis renderer)
panel (JFreeChart. plot)
frame (ChartFrame. "Histogram" panel)]
(doto plot
(.setBackgroundAlpha 0.0)
(.setRangeGridlinesVisible false)
(.setDomainGridlinesVisible false))
(doto renderer
(.setBarPainter painter)
(.setPaint blue)
(.setDrawBarOutline true)
(.setOutlinePaint blue)
(.setOutlineStroke (java.awt.BasicStroke. 1))
(.setShadowVisible false))
(doseq [[x y] values]
(.add series (+ x 0.5) y))
(.setLowerBound y-axis 0.5)
(.setVisible (.getLegend panel) false)
(doto frame
(.setSize 800 250)
(.setVisible true))))
I will omit a detailed explanation of how draw-hist works because I’m
still not a JFreeChart expert (though if I do many more blog posts on
this topic I may be forced to become one, however reluctantly). The
principal difference with the Incanter implementation of histograms is
that we provide our own set of bin heights and simply plot those. This
follows a better separation of concerns anyways: our fairly simple
binning function remains separate from the visual presentation of the
bin positions and heights (we could, for example, add the counting of
overflows and underflows – this is left as an exercise to the reader).
(trim-zeros exists to transform bin values of zero to small positive
numbers to avoid taking the logarithm of zero, which is undefined.)
Armed with these tools, we can now make our first distribution:
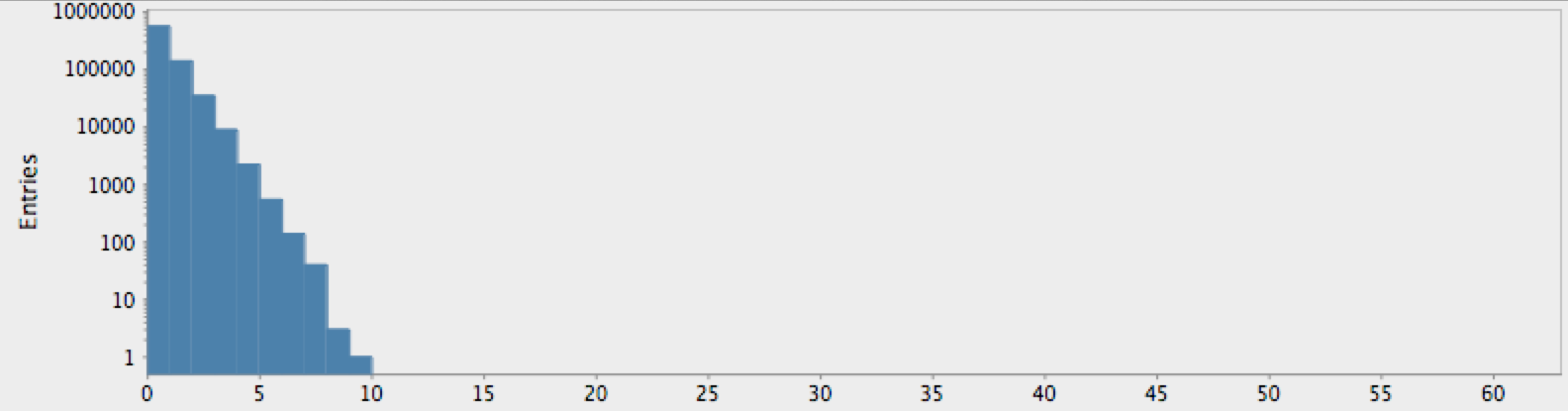
(->> (randgenome)
(take 1000000)
get-lengths
(make-hist 0.5 60.5 60)
trim-zeros
(draw-hist "Repeat Lengths, random hypothesis"))
As expected by our analysis above, this shows the falling “power-law” distribution of randomly-occuring nucleotide repetitions. Compare this with the genome for the yeast S. Cerviciae:
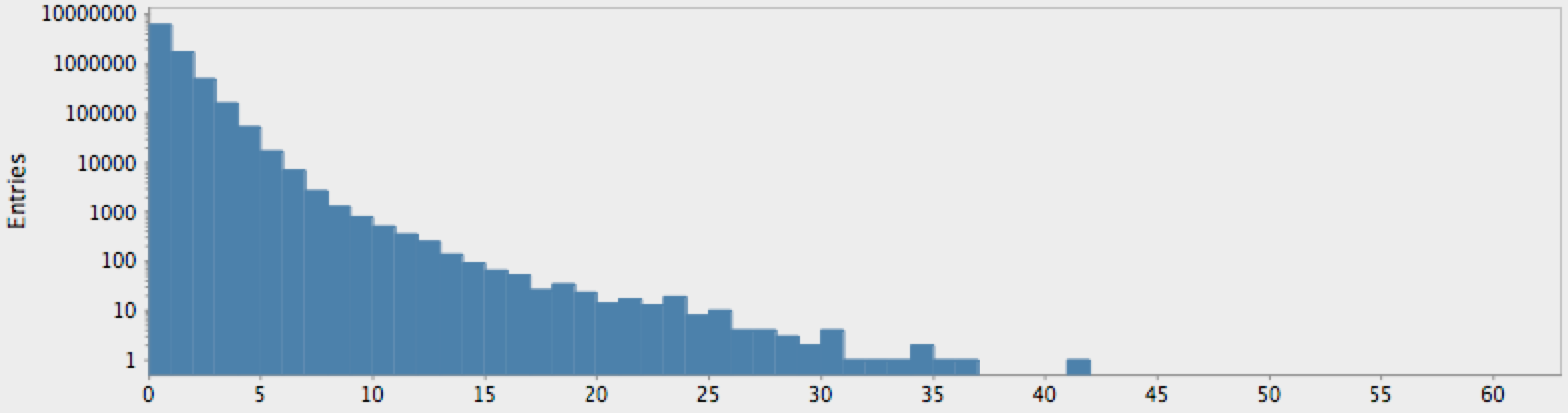
(->> (genome-sequence yeast)
get-lengths
(make-hist 0.5 60.5 60)
trim-zeros
(draw-hist "Repeat Lengths, S. Cerviciae"))
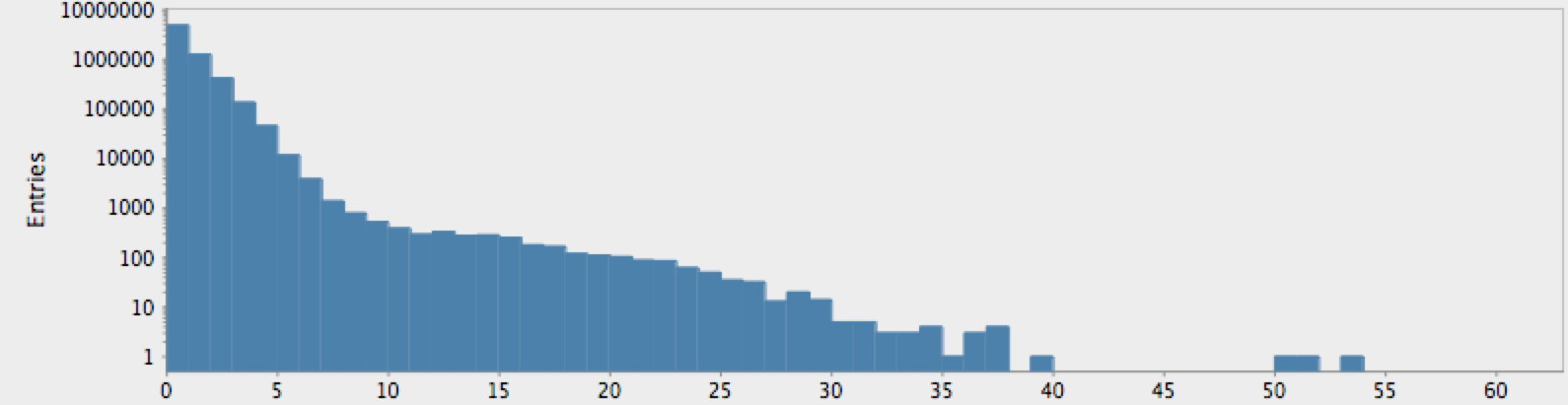
And, for humans (here we did not wait for the result for the entire genome):
(->> (genome-sequence human)
(take 10000000)
get-lengths
(make-hist 0.5 60.5 60)
trim-zeros
(draw-hist "Repeat Lengths, human genome"))
The actual genome data clearly deviate from our null hypothesis of randomness, as one might expect. However, these graphs raise more questions than they answer. In particular, note that there is a sort of bimodal characteristic or extended, secondary bump in the data for humans, with hints of an outlier feature on the tail which may or may not just be a statistical fluctuation.
One complicating factor we neglected is the role of “N-blocks”, which
should be eliminated without introducing artificially longer lengths;
e.g. ANNNNA with the Ns removed should be considered two 1-blocks
rather than a two-block AA group. This simple modification to
get-lengths is left as another exercise to the reader (the change does
not affect the resulting distributions).
A real biologist could no doubt tell us a lot about these distributions. Meanwhile, the latest code has been pushed to GitHub.
Blog Posts (170)
Select from below, view all posts, or choose only posts for:art clojure code emacs lisp misc orgmode physics python ruby sketchup southpole writing
From Elegance to Speed code lisp clojure physics Wednesday, September 25, 2019
Common Lisp How-Tos code lisp Sunday, September 15, 2019
Implementing Scheme in Python code python lisp Friday, September 13, 2019
A Daily Journal in Org Mode writing code emacs Monday, September 2, 2019
Show at Northwestern University Prosthetics-Orthotics Center art Saturday, October 20, 2018
Color Notations art Thursday, September 20, 2018
Painting and Time art Tuesday, May 29, 2018
Learning Muscular Anatomy code clojure art emacs orgmode Thursday, February 22, 2018
Reflections on a Year of Daily Memory Drawings art Monday, September 18, 2017
Repainting art Sunday, May 14, 2017
Daily Memory Drawings art Tuesday, January 3, 2017
Questions to Ask art Thursday, February 25, 2016
Macro-writing Macros code clojure Wednesday, November 25, 2015
Time Limits art Saturday, April 25, 2015
Lazy Physics code clojure physics Thursday, February 12, 2015
Fun with Instaparse code clojure Tuesday, November 12, 2013
Updating the Genome Decoder code clojure Saturday, July 13, 2013
Getting Our Hands Dirty (with the Human Genome) code clojure Wednesday, July 10, 2013
Validating the Genome Decoder code clojure Sunday, July 7, 2013
A Two Bit Decoder code clojure Saturday, July 6, 2013
Exploratory Genomics with Clojure code clojure Friday, July 5, 2013
Rosalind Problems in Clojure code clojure Sunday, June 9, 2013
Introduction to Context Managers in Python code python Saturday, April 20, 2013
Processes vs. Threads for Integration Testing code python Friday, April 19, 2013
Resources for Learning Clojure code clojure Monday, May 21, 2012
Continuous Testing in Python, Clojure and Blub code clojure python Saturday, March 31, 2012
Programming Languages code clojure python Thursday, December 22, 2011
Milvans and Container Malls southpole Friday, December 2, 2011
Oxygen southpole Tuesday, November 29, 2011
Ghost southpole Monday, November 28, 2011
Turkey, Stuffing, Eclipse southpole Sunday, November 27, 2011
Wind Storm and Moon Dust southpole Friday, November 25, 2011
Shower Instructions southpole Wednesday, November 23, 2011
Fresh Air and Bananas southpole Sunday, November 20, 2011
Traveller and the Human Chain southpole Thursday, November 17, 2011
Reveille southpole Monday, November 14, 2011
Drifts southpole Sunday, November 13, 2011
Bon Voyage southpole Friday, November 11, 2011
A Nicer Guy? southpole Friday, November 11, 2011
The Quiet Earth southpole Monday, November 7, 2011
Ten southpole Tuesday, November 1, 2011
The Wheel art Wednesday, October 19, 2011
Plein Air art Saturday, August 27, 2011
ISO50 southpole art Thursday, July 28, 2011
SketchUp and MakeHuman sketchup art Monday, June 27, 2011
In Defense of Hobbies misc code art Sunday, May 29, 2011
Closure southpole Tuesday, January 25, 2011
Takeoff southpole Sunday, January 23, 2011
Mummification southpole Sunday, January 23, 2011
Eleventh Hour southpole Saturday, January 22, 2011
Diamond southpole Thursday, January 20, 2011
Baby, It's Cold Outside southpole Wednesday, January 19, 2011
Fruition southpole Tuesday, January 18, 2011
Friendly Radiation southpole Tuesday, January 18, 2011
A Place That Wants You Dead southpole Monday, January 17, 2011
Marathon southpole Sunday, January 16, 2011
Deep Fried Macaroni and Cheese Balls southpole Saturday, January 15, 2011
Retrograde southpole Thursday, January 13, 2011
Three southpole Wednesday, January 12, 2011
Transitions southpole Tuesday, January 11, 2011
The Future southpole Monday, January 10, 2011
Sunday southpole Sunday, January 9, 2011
Radio Waves southpole Saturday, January 8, 2011
Settling In southpole Thursday, January 6, 2011
Revolution Number Nine southpole Thursday, January 6, 2011
Red Eye to McMurdo southpole Wednesday, January 5, 2011
That's the Way southpole Tuesday, January 4, 2011
Faults in Ice and Rock southpole Monday, January 3, 2011
Bardo southpole Monday, January 3, 2011
Chasing the Sun southpole Sunday, January 2, 2011
Downhole southpole Monday, December 13, 2010
Coming Out of Hibernation southpole Monday, September 13, 2010
Managing the Most Remote Data Center in the World code southpole Friday, July 30, 2010
Ruby Plugins for Sketchup art sketchup ruby code Saturday, April 3, 2010
The Cruel Stranger misc Tuesday, November 10, 2009
Photoshop on a Dime art Monday, October 12, 2009
Man on Wire misc Friday, October 9, 2009
Videos southpole Friday, May 1, 2009
Posing Rigs art Sunday, April 5, 2009
Metric art Monday, March 2, 2009
Cuba southpole Sunday, February 22, 2009
Wickets southpole Monday, February 16, 2009
Safe southpole Sunday, February 15, 2009
Broken Glasses southpole Thursday, February 12, 2009
End of the Second Act southpole Friday, February 6, 2009
Pigs and Fish southpole Wednesday, February 4, 2009
Last Arrivals southpole Saturday, January 31, 2009
Lily White southpole Saturday, January 31, 2009
In a Dry and Waterless Place southpole Friday, January 30, 2009
Immortality southpole Thursday, January 29, 2009
Routine southpole Thursday, January 29, 2009
Tourists southpole Wednesday, January 28, 2009
Passing Notes southpole Thursday, January 22, 2009
Translation southpole Tuesday, January 20, 2009
RNZAF southpole Monday, January 19, 2009
The Usual Delays southpole Monday, January 19, 2009
CHC southpole Saturday, January 17, 2009
Wyeth on Another Planet art Saturday, January 17, 2009
Detox southpole Friday, January 16, 2009
Packing southpole Tuesday, January 13, 2009
Nails southpole Friday, January 9, 2009
Gearing Up southpole Tuesday, January 6, 2009
Gouache, and a new system for conquering the world art Sunday, November 30, 2008
Fall 2008 HPAC Studies art Friday, November 21, 2008
YABP (Yet Another Blog Platform) southpole Thursday, November 20, 2008
A Bath southpole Monday, February 18, 2008
Green Marathon southpole Saturday, February 16, 2008
Sprung southpole Friday, February 15, 2008
Outta Here southpole Wednesday, February 13, 2008
Lame Duck DAQer southpole Tuesday, February 12, 2008
Eclipse Town southpole Saturday, February 9, 2008
One More Week southpole Wednesday, February 6, 2008
IceCube Laboratory Video Tour; Midrats Finale southpole Friday, February 1, 2008
SPIFF, Party, Shift Change southpole Monday, January 28, 2008
Good things come in threes, or 18s southpole Saturday, January 26, 2008
Sleepless in the Station southpole Thursday, January 24, 2008
Post Deploy southpole Monday, January 21, 2008
IceCube and The Beatles southpole Saturday, January 19, 2008
Midrats southpole Saturday, January 19, 2008
Video: Flight to South Pole southpole Thursday, January 17, 2008
Almost There southpole Wednesday, January 16, 2008
The Pure Land southpole Wednesday, January 16, 2008
There are no mice in the Hotel California Bunkroom southpole Sunday, January 13, 2008
Short Timer southpole Sunday, January 13, 2008
Sleepy in MacTown southpole Saturday, January 12, 2008
Sir Ed southpole Friday, January 11, 2008
Pynchon, Redux southpole Friday, January 11, 2008
Superposition of Luggage States southpole Friday, January 11, 2008
Shortcut to Toast southpole Friday, January 11, 2008
Flights: Round 1 southpole Thursday, January 10, 2008
Packing for the Pole southpole Monday, January 7, 2008
Goals for Trip southpole Sunday, January 6, 2008
Balaklavas southpole Friday, January 4, 2008
Tree and Man (Test Post) southpole Friday, December 28, 2007
Schedule southpole Sunday, December 16, 2007
How to mail stuff to John at the South Pole southpole Sunday, November 25, 2007
Summer and Winter southpole Tuesday, March 6, 2007
Homeward Bound southpole Thursday, February 22, 2007
Redeployment southpole Monday, February 19, 2007
Short-timer southpole Sunday, February 18, 2007
The Cleanest Air in the World southpole Saturday, February 17, 2007
One more day (?) southpole Friday, February 16, 2007
One more week (?) southpole Thursday, February 15, 2007
Closing Softly southpole Monday, February 12, 2007
More Photos southpole Friday, February 9, 2007
Super Bowl Wednesday southpole Thursday, February 8, 2007
Night Owls southpole Tuesday, February 6, 2007
First Week southpole Friday, February 2, 2007
More Ice Pix southpole Wednesday, January 31, 2007
Settling In southpole Tuesday, January 30, 2007
NPX southpole Monday, January 29, 2007
Pole Bound southpole Sunday, January 28, 2007
Bad Dirt southpole Saturday, January 27, 2007
The Last Laugh southpole Friday, January 26, 2007
First Delay southpole Thursday, January 25, 2007
Nope southpole Thursday, January 25, 2007
Batteries and Sheep southpole Wednesday, January 24, 2007
All for McNaught southpole Tuesday, January 23, 2007
t=0 southpole Monday, January 22, 2007
The Big (Really really big...) Picture southpole Monday, January 22, 2007
Giacometti southpole Monday, January 22, 2007
Descent southpole Monday, January 22, 2007
Video Tour southpole Saturday, January 20, 2007
How to subscribe to blog updates southpole Monday, December 11, 2006
What The Blog is For southpole Sunday, December 10, 2006
Auckland southpole Tuesday, January 11, 2005
Halfway Around the World; Dragging the Soul Behind southpole Monday, January 10, 2005
Launched southpole Sunday, January 9, 2005
Getting Ready (t minus 2 days) southpole Friday, January 7, 2005
Subscribe: RSS feed ... all topics ... or Clojure only / Lisp only